1. Introduction
hloroplasts, the organelles responsible for photosynthesis, are in many respects similar to mitochondria.
Both chloroplasts and mitochondria function to generate metabolic energy, evolved by endosymbiosis, contain their own genetic systems, and replicate by division. However, chloroplasts are larger and more complex than mitochondria, and they perform several critical tasks in addition to the generation of ATP. Most importantly, chloroplasts are responsible for the photosynthetic conversion of Carbon Di-oxide to carbohydrates. In addition, chloroplasts synthesize amino acids, fatty acids, and the lipid components of their own membranes. The reduction of nitrite to ammonia, an essential step in the incorporation of nitrogen into organic compounds, also occurs in chloroplasts. Moreover, chloroplasts are only one of several types of related organelles (plastids) that play a variety of roles in plant cells [1][2][3][4][5][6][7].
Microsatellites (sometimes referred to as a variable number of tandem repeats or VNTRs) are short segments of DNA that have a repeated sequence, and they tend to occur in DNA. In some microsatellites, the repeated unit may occur four times, in others it may be seven, or two, or three [8]. These repeats are ubiquitous in nature and are responsible for causing several diseases and cancers [9] [10].
These are used in various applications like DNA Fingerprinting, DNA Forensics, Paternity Studies, and have been considered as potential markers for identifying species, for establishing phylogenetic relationships and also to study evolution [11]. Microsatellites are ubiquitously found in both coding and non-coding regions of all organisms and their distribution in coding regions (genes) is known to affect protein formation and gene regulation [12].
Next-generation sequencing enabled researchers to study biological systems at a level never before possible. Studying mutations in chloroplast microsatellite repeats can be very helpful to understand various biological questions and their usage in various other diverse applications. Few studies [13][14][15][16] earlier analyzed the distribution of microsatellites in chloroplast genomes but they are only confined to single or very low number of genomes. This paper describes the study performed to analyze microsatellite repeats in more than 370 chloroplasts genomes and details have been presented.
2. II.
3. Materials & Methods
Imperfect microsatellites have been extracted from Chloro Mito SSRDB [17] version 2.0, an opensource microsatellite repository of sequenced organelle genomes. For this study, a total of 370 chloroplast genome sequences have been used that belong to various classes as shown in Table 1.
4. Discussion a) Genome Size Analysis
We did a preliminary study to analyze the genome sizes of all chloroplasts. The chloroplast genome sizes vary from few kbs to a maximum of 1 Mb. The smallest chloroplast genome reported is of size 29529bp that belongs to plant named Plasmodium falciparum HB3 apicoplast (ID: NC_017928) belongs to Non-Viridiplantae category. The largest chloroplast genome spans about 1021616 bp of length that belongs to Paulinella chromatophora chromatophore (ID: NC_011087) belongs to Rhizaria.
In Viridiplantae, the smallest chloroplast genome is Helicosporidium sp. ex Simulium jonesii plastid(ID: NC_008100) of length 37454 bp where as the largest chloroplast genome is Floydiella terrestris(ID: NC_014346) chloroplast of length 521168 bp.
In Non-Viridiplantae, the smallest chloroplast genome is found as Plasmodium falciparum HB3 apicoplast (ID: of length 29529 bp where as the largest chloroplast genome is Paulinella chromatophora chromatophore (ID: NC_011087) chloroplast of length 1021616 bp. It is observed that this non-Virdiplantae category genome size is greater than the Viridiplantae genomes.
When the average genome sizes of chloroplast are considered category wise, it has been observed that the average lengths of Viridiplantae chloroplast genomes are little bit higher when compared to those of other non Virdiplantae(Refer Fig 1). 2 gives a summary of the total number of genomes categorized based on genome sizes of the two classes of chloroplast. It has been observed that majority of the genome sizes lie between 10kb to 500kb, only two genomes namely Floydiella terrestris chloroplast (NC_014346) and Paulinella chromatophora chromatophore (NC_011087) are found to be greater than 500kb. On the other hand, 311 plants of Viridiplantae show genome sizes between 100kb and 500kb.
5. b) Distribution of Microsatellites
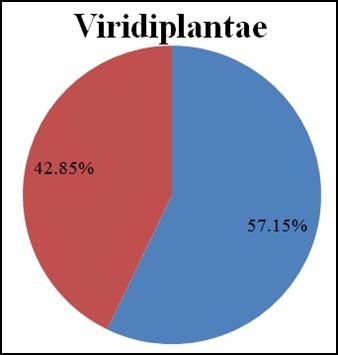
Microsatellites in or near genes (coding regions) are found to impact protein formation and gene regulation. When the distribution of microsatellites has been analyzed overall, it is found that around 57% of microsatellite repeats fall in coding regions of all chloroplast genomes. Out of the total 78536 chloroplast microsatellites, 45518 microsatellites fall in gene regions where as the rest 33018 repeats fall in non-coding regions. However, it is surprising to see that the distribution differs when the two classes have been compared separately (Refer Fig. 2). Genomes of Non-Viridiplantae are found to be having majority of its microsatellites in coding regions (64%). On the other hand, green plants (Viridiplantae) show that around 57% of their microsatellites to be distributed in coding regions. When two chloroplast categories are compared (Refer Fig. 3), these two categories exhibit a similar distribution of its microsatellites in coding and non coding regions. It would be interesting to study the reason behind the major number of microsatellite repeats in Viridiplantae.
6. c) Motif-size wise Analysis
We have further analyzed the distribution of chloroplast microsatellites based on their motif sizes. Table 3 lists the proportionate distribution of chloroplast microsatellites motif-size wise. It has been observed that chloroplast genomes are rich in tri and tetra nucleotide repeats which tohether account for more than 77% in Non-virdiplantae, and around 62% in Virdiplantae. Mono, Penta and Hexa-nucleotide repeats are found to be very low in number. When the microsatellite tract lengths have been analyzed, the genomes reported few interesting tract lengths for almost all motif sizes. The average microsatellite tract lengths are usually observed to be not more than 19 bp. But, it is surprising to note that some of the tetra and tri repeats have shown exceptional tract lengths as large as 276bp have been observed. Based on the results in Table 4, we have further tried to find repeats in chloroplast genomes that have exceptional tract lengths. Interestingly, we found 10 repeats in chloroplast with tract lengths 100bp or more; out of those, two repeats have tract lengths 200bp or more. Two significant tract lengths of 276 and 203 have been reported for genomes with IDs NC_020321, NC_008117 respectively.
IV.
7. Conclusion
In this paper, we have presented a brief description about the distribution of microsatellite repeats in all sequenced chloroplast genomes of Plants. This study forms the first comprehensive analysis of microsatellite repeats in chloroplast genomes and the statistics of this study can be a useful resource for biologists.


| Category | Total No. | ||
| Alveolata | 9 | ||
| Cryptophyta | 3 | ||
| Euglenozoa | 5 | ||
| Glaucocystophyceae | 1 | ||
| Haptophyceae | 4 | ||
| Rhizaria | 2 | ||
| Rhodophyta | 9 | ||
| Stramenopiles | 14 | ||
| Viridiplantae | 323 | ||
| Total Genomes | 370 | ||
| Among the 370 genomes, 323 genomes belong | |||
| to Viridiplantae (Green Plants), 47 genomes belongs to | |||
| Non-Viridiplantae which include genomes of Alveolata, | |||
| Cryptophyta, | Euglenozoa, | Glaucocystophyceae, | |
| Haptophyceae, | Rhizaria, | Rhodophyta | and |
| Average genome sizes of chloroplast | ||||
| 150000 | 148178.28 | |||
| 145000 | ||||
| Genome size | 140000 | 136551.53 | ||
| 135000 | ||||
| 130000 | ||||
| Viridiplantae | Non-Viridiplantae | |||
| Size Range | No. of plants | |||
| >= 10 Kb and <50 Kb | ||||
| Non-Viridiplantae | 5 | |||
| Viridiplantae | 2 | |||
| >= 50 Kb and <100 Kb | ||||
| Non-Viridiplantae | 10 | |||
| Viridiplantae | 9 | |||
| >= 100 Kb and <500 Kb | ||||
| Non-Viridiplantae | 31 | |||
| Motif Size | Non-Viridiplantae | Viridiplantae |
| Mono | 159(1.80%) | 8602(12.33%) |
| Di | 840(9.55%) | 7909(11.34%) |
| Tri | 3506(39.87%) | 17055(24.45%) |
| Tetra | 3300(37.52) | 26796(38.42%) |
| Penta | 623(7.08%) | 5680(8.14%) |
| Hexa | 365(4.15%) | 3701(5.31%) |
| Total | 8793 | 69743 |
| Non-Viridiplantae | Viridiplantae | |||||
| Motif Size | High | Low | Avg | High | Low | Avg |
| MONO | 25 | 12 | 13.93 | 46 | 12 | 14.49 |
| DI | 54 | 11 | 12.90 | 83 | 11 | 13.24 |
| TRI | 51 | 11 | 12.19 | 276 | 11 | 12.38 |
| TETRA | 29 | 11 | 11.91 | 203 | 11 | 12.13 |
| PENTA | 65 | 14 | 15.27 | 100 | 14 | 15.41 |
| HEXA | 42 | 17 | 18.74 | 145 | 17 | 19.70 |