1. On the Notion of Percentage Nucleotide Concentration of Genome Sequences in Terms of Cellular Automata Evolutions of Adjoints Sequences
Prashanthi Govindarajan ? , Sathya Govindarajan ? & Ethirajan Govindarajan ?
Abstract-This paper proposes a novel concept called "Percentage Nucleotide Concentration of genomes" in terms of cellular automata evolutions of adjoints of Adenine, Thymine, Guanine, and Cytosine. The adjoints of the given a genome sequenceare the characteristic binary string sequences. For example, the adjoint of Adenine of a given genome sequence is a binary string consisting of 0's and 1's where 1's corresponds to the presence of Adenine in the genome sequence. So, one can have four adjoint sequences of Adenine, Thymine, Guanine, and Cytosine corresponding to a given genome sequence. One-dimensional three neighborhood binary value cellular automata rules could be applied to an adjoint sequence and the desired number of evolutions obtained.These rules aredefined by linear Boolean functions and one can have 256 such linear Boolean functions.Nucleotide concentration is computed for an adjoint sequence and its variation evaluated for its successive evolutions based on a cellular automaton rule.
2. I. Introduction
he purpose of the research carried out and reported in this paper is whether it is possible to categorize a set of genomes like the human genome repository. The concept of "%nucleotide concentration" introduced in this paper seems to show a way to accomplish this task. The genesis of the formulation of this concept originates from chemistry, wherein the quantificational notion of percentage ionic concentration of hydrogen (pH value) is used to categorize solutions into three (i) water, whose pH value is 7, (ii) acidic solutions whose pH values are less than 7 and (iii) alkaline solutions whose pH values are greater than 7. On the same lines, an effort was made to categorize genome sets based on four values (i) % nucleotide concentration of Adenine (pA), (ii) % nucleotide concentration of Thymine (pT), (iii) % nucleotide concentration of Guanine (pG) and (iv) % nucleotide concentration of Cytosine (pC). It is reasonable to surmise that these values, possibly their compositions would categorize a given set of genomes. The formulation of the concept is briefly explained below. Section 2 of this paper describes the concept formulation.
Section 3 of this paper describes the fundamental notions of adjoints of a genome and their evolution using one dimensional cellular automata rules defined by linear Boolean functions. Section 4 provides experimental results of a case study pertaining to evaluation of Concentration of Nucleotides in terms of Adjoints of BrucellaSuis 1330 Genome Sequence.
3. II. Concept Formulation
Analogous to the notion of pH value of a solution, the values of pA, pT, pG and pC of a genome sequence and possibly composition of these values like the proportion pA:pT:pG:pC seems to pave a way to classify and characterize genome sets. The definition of "Percentage Nucleotide Concentration" of a genome sequence is given below.
4. Definition
Given a genome sequence, the number of a particular nucleotide, say A, present in that genome sequence is counted and the sum is divided by the total number of nucleotides in that genome sequence. The fraction when multiplied by 100 yields the "Percentage Concentration of Adenine pA". Similarly, one can evaluate pT, pG and pC.
5. One-Dimensional Three Neighborhood Cellular Automata Evolutions of Adjointsof a Genome Sequence
Adjoint of a particular nucleotide in a genome sequence is the binary sequence obtained by substituting the particular nucleotides in the genome sequence by 1's and the others by 0's. For example, let us consider a sample sequence of BrucellaSuis 1330 for a case study. The actual length of the genome sequence of BrucellaSuis 1330 is 5806. A cellular T automaton is an idealized parallel processing system consisting of an array of numbers (1-D, 2-D and more) realized using updating rules based on certain neighborhood. For example, a one-dimensional cellular automaton would consist of a finite-length array as shown below.
--- --- --- i-1 i i+1 --- --- ---Consider an ith cell in the array. This cell has a neighbor i-1 on its left and another i+1 on its right. All three put together is called a three neighborhood. One can assign a site (cell) variable ?i-1, ?i, and ?i+1 to the three neighborhood cells. At a particular instant of time, these variables take on numerical values, say either a 0 or a 1. In such a case, the variables are denoted as ?ti-1, ?ti, and ?ti+1. The value of the ith cell at the next instant of time is evaluated using an updating rule that involves the present values of the ith, (i-1)th and (i+1)th cells. This updating rule is essentially a linear Boolean function of three variables. One can construct 256 linear Boolean functions as updating rules of one-dimensional threeneighborhood binary-valued cellular automata. Each rule defines an automaton by itself. So, one dimensional binary valued three neighborhood cellular automata (123CA) rules could be used to model adjoints of a genome sequence. The first twenty linear Boolean functions of cellular automata 123CA are listed below with their decimal equivalents.
6. Linear Boolean Function
For the case study rule number 90 is applied to the adjoints of BrucellaSuis 1330 genome sequence and 500 evolutions generated. Rule 90 is shown below.
(? ???1 ? ? ??+1 ) + (? ? ???1 ? ??+1 )90Since the image of the 500 evolutions of BrucellaSuis 1330 is large, a small portion of the images are presented in this paper.
7. Concentration of Nucleotidesin Adjoints of Brucellasuis 1330 Genome Sequence
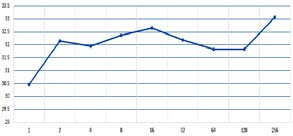
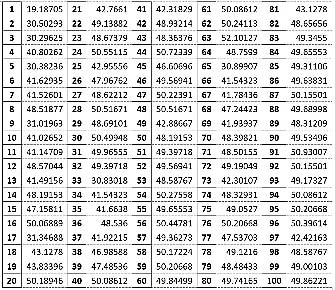
The values of pA, pT, pG and pCof the BrucellaSuis 1330 genome sequence are computed for the adjoints A(n), T(n),G(n) and C(n) and their 500 evolutions using 123CA rules based one linear Boolean functions. Fig. 1 shows the evolutions of the adjoints of A(n), T(n). G(n) and C(n) using the linear Boolean function rule 90 of 123CA. The values are tabulated and the corresponding graphs shown subsequently. Table 1 shows the pA values of A(n) of BrucellaSuis 1330 genome sequence and the 500 generations of A(n) using rule 90 of 123CA. Figs. 2 and 3 shows the graphs of the variations of pA values of all generations. Table 2 shows the pT values of T(n) of BrucellaSuis 1330 genome sequence and the 500 generations of T(n) using rule 90 of 123CA. Figs. 4and 5 shows the graph of the variations of pT values of all generations. Table 3 shows the pG values of G(n) of BrucellaSuis 1330 genome sequence and the 500 generations of G(n) using rule 90 of 123CA. Fig. 4 shows the graph of variations of pA values of all generations. Table 4 shows the pC values of C(n) of BrucellaSuis 1330 genome sequence and 500 generations of C(n) using rule 90 of 123CA. Fig. 5 shows the graph of the variations of pC values of all generations. This paper proposes a novel concept called "Percentage Nucleotide Concentration of genomes" in terms of cellular automata evolutions of adjoints of Adenine, Thymine, Guanine, and Cytosine. The research carried out and reported in this paper exhibits the possibility to categorize a set of genomes like the human genome repository. In short, the concept of "Percentage Nucleotide Concentration (PNC)" introduced in this paper seems to show a way to accomplish this task.


| Rule number 90 is applied to A(n) | |||
| and its 500 generations. It is | |||
| observed that the pA value | |||
| becomes minimum at regular | |||
| intervals of 1, 2, 4, 8, 16, 32, 64, 128 | |||
| and 256. This indicates a fractal | |||
| behavior | of | the | evolution. |
| Min(A(n))=30.2965 | and | ||
| Max(A(n))=31.4502. The deviation is | |||
| 1.15. | |||
| Rule number 90 is applied to | |
| T(n) and its 500 generations. It | |
| is observed that the pT value | |
| becomes minimum at regular | |
| intervals of 1, 2, 4, 8, 16, 32, | |
| 64, 128 and 256. This | |
| indicates a fractal behavior of | |
| the evolution. | |
| Min(A(n))=30.45126; | |
| Max(A(n))=33.06924. | The |
| deviation is 2.61. | |
| Rule number 90 is applied to G(n) | |||
| and its 500 generations. It is | |||
| observed that the pG value | |||
| becomes minimum at regular | |||
| intervals of 1, 2, 4, 8, 16, 32, 64, 128 | |||
| and 256. This indicates a fractal | |||
| behavior | of | the | evolution. |
| Min(A(n))=43.00723 | and | ||
| Max(A(n))=44.29900 The deviation | |||
| is 1.46. | |||