1. Modeling and Simulation of Genome Evolution Using Linear Boolean Functions Associated with
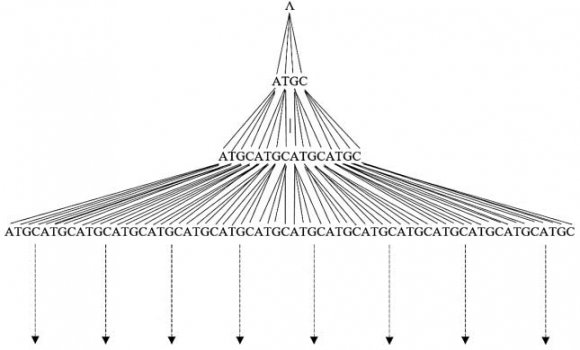
One Dimensional Cellular Automata Prashanthi Govindarajan ? , Sathya Govindarajan ? & Ethirajan Govindarajan ? I. Introduction To be precise, Fig. 1 shows three levels of nucleotides. One can generate 64 strands of length 3. As the length increases, the number of strands he four nucleotides A, T, G, and C get connected by phosphodiester bonds to form strands. Strand formation depends on innumerable factors related to inter and intra cellular parameters and functions. One cannot precisely say that a particular strand gets formed using such and such rules. The infinite possibilities of strand formation cannot be determined experimentally or in the framework of classical genetics. One can alternatively formulate a notion of "Language of Genomes" wherein one can finitely specify infinite strands, Fig. 1 shows a finitely generated quaternary tree structure of strand formation of nucleic acids. T increases as per the formula 4n, where n is the length of the strand. Strands of length three are called triplet codons or 3-tuple codons. Similarly, one can think of ntuple codons where n is any number.
A genome sequence is a chain of four nucleotides A, T, G and C. The numerical representation of a genome sequence is a sequence of four numbers 1, 2, 3 and 4. Linear prediction of a strand could be carried out using linear prediction algorithms from a sub sequence of length 8. Alternatively, one can evolve generations of genome sequences from a given fulllength genome sequence using one-dimensional cellular automata rules. Section 2 describes the notions of adjoints of nucleotides corresponding to a genome sequence. Section 3 describes the notions of cellular automata and linear Boolean functions. Section 4 provides the results of applying linear Boolean functions on adjoint strings of nucleotides. Section 5 demonstrates the results of combining evolution patterns of adjoint sequences dyadically. Section 6 presents various observations made from the study and proposes future perspectives of cellular automatabased genome analytics.
Adjoint of a particular nucleotide in a genome sequence is the binary sequence obtained by substituting the particular nucleotides in the genome sequence by 1's and the others by 0's. For example, let us consider a sample sequence G, A, A, T, G, A, T, T, A, C, C, A, A, G, G, C of length 16. Now the adjoint of adenine (A) is the binary string A(n) = 0, 1, 1, 0, 0, 1, 0, 0, 1, 0, 0, 1, 1, 0, 0, 0. The adjoint of thymine (T) is the binary string T(n) = 0, 0, 0, 1, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0. The adjoint of guanine (G) is the binary string G(n) = 1, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 1, 1, 0, 0. The adjoint of cytocine (C) is binary string C(n) = 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 1, 0, 0, 0, 0, 1. The first segment of 40 nucleotides of a genome sequence of Brucella Suis 1330 is considered here for a case study. The actual length of the genome sequence of Brucella Suis 1330 is 5806. The sample sequence is given below.
A(n) = 0110010010011000011000011000000000000000 T(n) = 0001001100000000000001000001010011001100 G(n) = 1000100000000110000100000100000000000011 C(n) = 0000000001100001100010100010101100110000A cellular automaton is an idealized parallel processing system consisting of an array of numbers (1-D, 2-D and more) realized using updating rules based on certain neighborhood. For example, a onedimensional cellular automaton would consist of a finite length array as shown below.
2. III. Cellular Automata and Linear Boolean Functions
A cellular automaton is an idealized parallel processing system consisting of an array of numbers (1-D, 2-D and more) realized using updating rules based on certain neighborhood. For example, a one dimensional cellular automaton would consist of a finite length array as shown below.
-
-- --- --- i-1 i i+1 --- --- ---Consider an ith cell in the array. This cell has a neighbor i-1 on its left and another i+1 on its right. All three put together is called a three-neighborhood. One can assign a site (cell) variable ?i-1, ?i, and ?i+1 to the three-neighborhood cells. At a particular instant of time, these variables take on numerical values, say either a 0 or a 1. In such a case, the variables are denoted as ?ti-1, ?ti, and ?ti+1. The value of the ith cell at the next instant of time is evaluated using an updating rule that involves the present values of the ith, (i-1)th and (i+1)th cells. This updating rule is essentially a linear Boolean function of three variables. One can construct 256 linear Boolean functions as updating rules of one-dimensional threeneighborhood binary-valued cellular automata. Each rule defines an automaton by itself. So, one-dimensional binary-valued three-neighborhood cellular automata (123CA) rules could be used to model adjoints of a genome sequence. The first thirty linear Boolean functions of cellular automata 123CA are listed below with their decimal equivalents.
3. Linear Boolean Function
Decimal Equivalent 0 0
(?? ? ???1 ?? ? ?? ?? ? ??+1 ) 1 (?? ? ???1 ?? ? ?? ?? ??+1 ) 2 (?? ? ???1 ?? ? ?? ) 3 (?? ? ???1 ?? ?? ?? ? ??+1 ) 4 (?? ? ???1 ?? ? ??+1 ) 5 (?? ? ???1 ?? ?? ?? ? ??+1 )+(?? ? ???1 ?? ? ?? ?? ??+1 ) 6 (?? ? ???1 ?? ? ??+1 ) + (?? ? ???1 ?? ? ?? ) 7 (?? ? ???1 ?? ?? ?? ??+1 ) 8 (?? ? ???1 ?? ? ?? ?? ? ??+1 ) + (?? ? ???1 ?? ?? ?? ??+1 ) 9 (?? ? ???1 ?? ??+1 )10(?? ? ???1 ?? ? ?? ) + (?? ? ???1 ?? ??+1 ) (?? ? ???1 ?? ?? ) (?? ? ???1 ?? ? ??+1 ) + (?? ? ???1 ?? ?? ) (?? ? ???1 ?? ?? ) + (?? ? ???1 ?? ??+1 ) (?? ? ???1 ) (?? ???1 ?? ? ?? ?? ? ??+1 ) (?? ? ?? ?? ? ??+1 ) (?? ???1 ?? ? ?? ?? ? ??+1 ) + (?? ? ???1 ?? ? ?? ?? ??+1 ) (?? ? ?? ?? ? ??+1 ) + (?? ? ???1 ?? ? ?? ) (?? ???1 ?? ? ?? ?? ? ??+1 ) + (?? ? ???1 ?? ?? ?? ? ??+1 ) (?? ? ?? ?? ? ??+1 ) + (?? ? ???1 ?? ? ??+1 ) (?? ???1 ?? ? ?? ?? ? ??+1 ) + (?? ? ???1 ?? ?? ?? ? ??+1 ) + (?? ? ???1 ?? ? ?? ?? ??+1 ) (?? ? ?? ?? ? ??+1 ) + (?? ? ???1 ?? ? ??+1 ) + (?? ? ???1 ?? ? ?? ) (?? ???1 ?? ? ?? ?? ? ??+1 ) + (?? ? ???1 ?? ?? ?? ??+1 ) (?? ? ???1 ?? ?? ?? ??+1 ) + (?? ? ?? ?? ? ??+1 ) (?? ???1 ?? ? ?? ?? ? ??+1 ) + (?? ? ???1 ?? ??+1 ) (?? ? ?? ?? ? ??+1 ) + (?? ? ???1 ?? ??+1 ) (?? ???1 ?? ? ?? ?? ? ??+1 ) + (?? ? ???1 ?? ?? ) (?? ? ?? ?? ? ??+1 ) + (?? ? ???1 ?? ?? ) (?? ???1 ?? ? ?? ?? ? ??+1 ) + (?? ? ???1 ?? ?? ) + (?? ? ???1 ?? ??+1 )IV. Cellular Automata Evolutions of Genome Adjoints
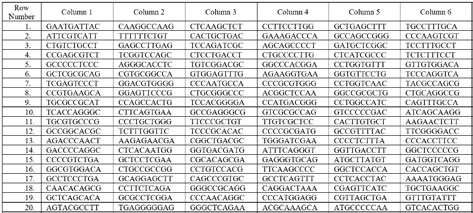
The genome sequence of Brucella Suis 1330 is considered here for a case study. Due to space limitations, a part of the genome sequence and its adjoints are shown below. As defined already, adjoint of genome sequence concerning a particular nucleotide is the binary string obtained by marking a '1' in the place of that particular nucleotide and by marking a '0' in the places of other nucleotides. A segment consisting of 60 nucleotides of Brucella Suis 1330 is shown below.
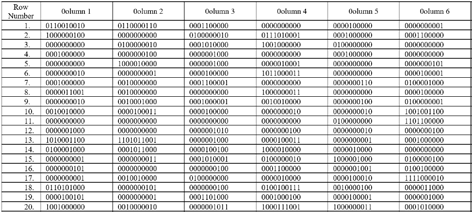
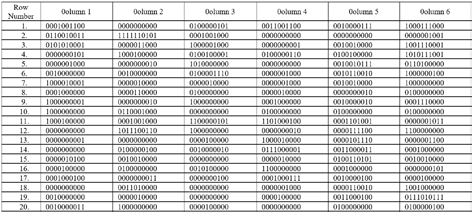
The adjoints of the genome sequence segment are given below.
Adjoint A(n)
Adjoint T(n) Adjoint G(n) Adjoint C(n)Cellular automata evolutions of adjoints of a genome are carried out using 256 rules of 123CA. As an example, rule number 137 of 123CA, that is, (? ? ???1 ? ? ?? ? ? ??+1 ) + (? ?? ? ??+1 ) is applied to adjoints of Brucella Suis 1330 genome and results shown below in Fig. 2.
Evolution of A(n) Evolution of T(n) Evolution of G(n) Evolution of C(n)Fig. 2: Evolution of adjoints using rule 137 of 123CA
The size of the images shown in Fig. 2 is 500x500, though the actual size is 5806x500. The first 500 columns of the actual images are clipped and presented here for visual clarity. From Fig. 2, it is clear that the evolution pattern of each adjoint is different. One can observe that there are certain fractal patterns in the evolutions and such fractals are distributed in the images very differently. For instance, the zoomed in versions of the evolution patterns of A(n), T(n), G(n) and C(n) using rule 137 are shown in Figs. 3, 4, 5 and 6 respectively.
4. VI. Observations and Conclusions
From the above empirical study, it is observed that cellular automata modeling and simulation of evolutions of adjoints of a given genome sequence and the inter-pattern operations and relations exhibit distinct patterns of fractals and fractal distributions. The novel technique and results presented in this paper are outcome of prolonged research carried out in the mathematical modeling of genomes and their evolutions. It is evident that one can as well look into the possibilities of genome editing using such cellular automata tools.